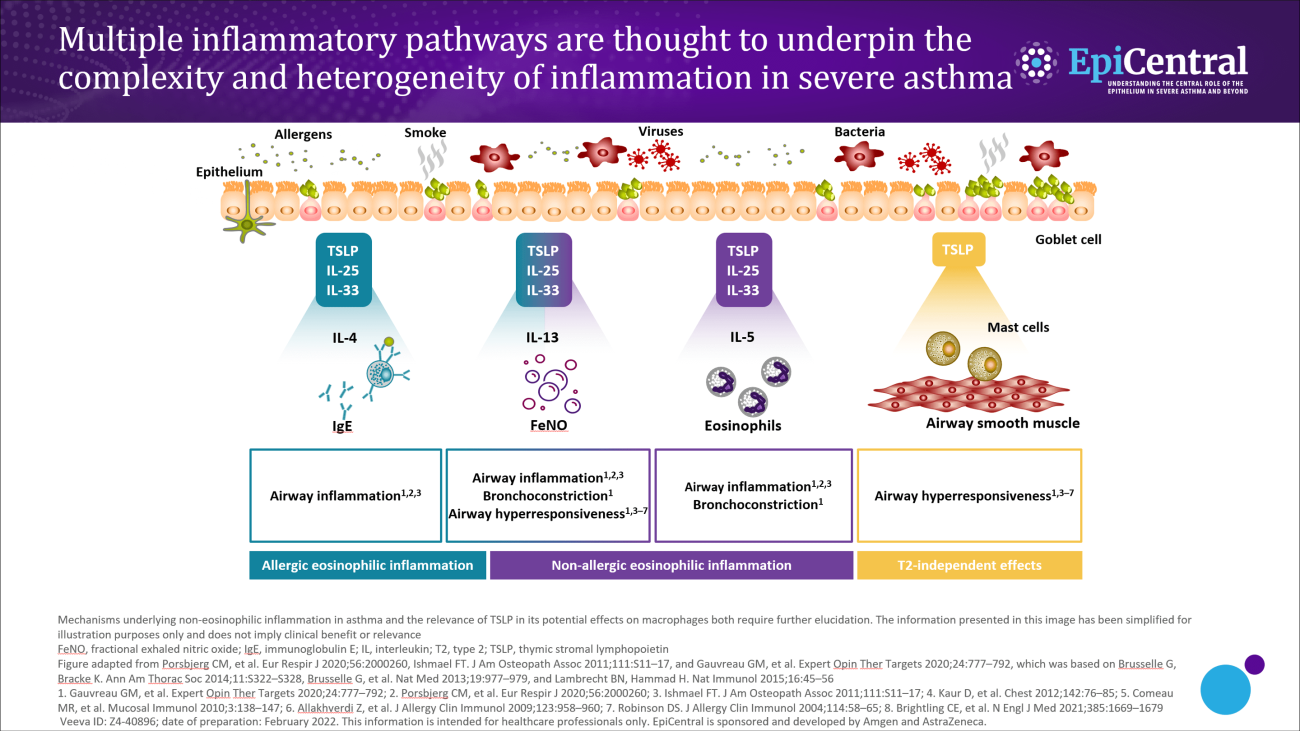
Some patients have concurrent overlapping inflammatory pathways.1–4 Often the extent of overlap is unknown; however, it is likely that many patients have a mixture of pathways that drive their disease.1,2 The dominant pathway may change over time owing to different circumstances, such as changes in medications, treatment adherence, exacerbations or exposure to allergens.1–4 In a biomarker study conducted in patients with severe asthma, the phenotype changed in ~50% of patients based on their sputum biomarker clustering after 1 year of follow-up.4
Upstream activation of epithelial cytokines can lead to initiation of multiple downstream inflammatory pathways.10 The wide spectrum and overlap of downstream pathways in severe asthma mean that diagnosis and treatment can be challenging.2 For example, in one US study, around a third of adults with severe asthma have both an allergic and an eosinophilic (defined as blood eosinophils >300 µL/cells) phenotype.2 It would be advantageous to further understand any overlap and common drivers (such as epithelial cytokines) of the downstream endotypes of severe asthma.11,12 Understanding this could help to identify appropriate treatments to address the overlap of downstream pathways and to improve patient care.6,11
Biomarkers in severe asthma
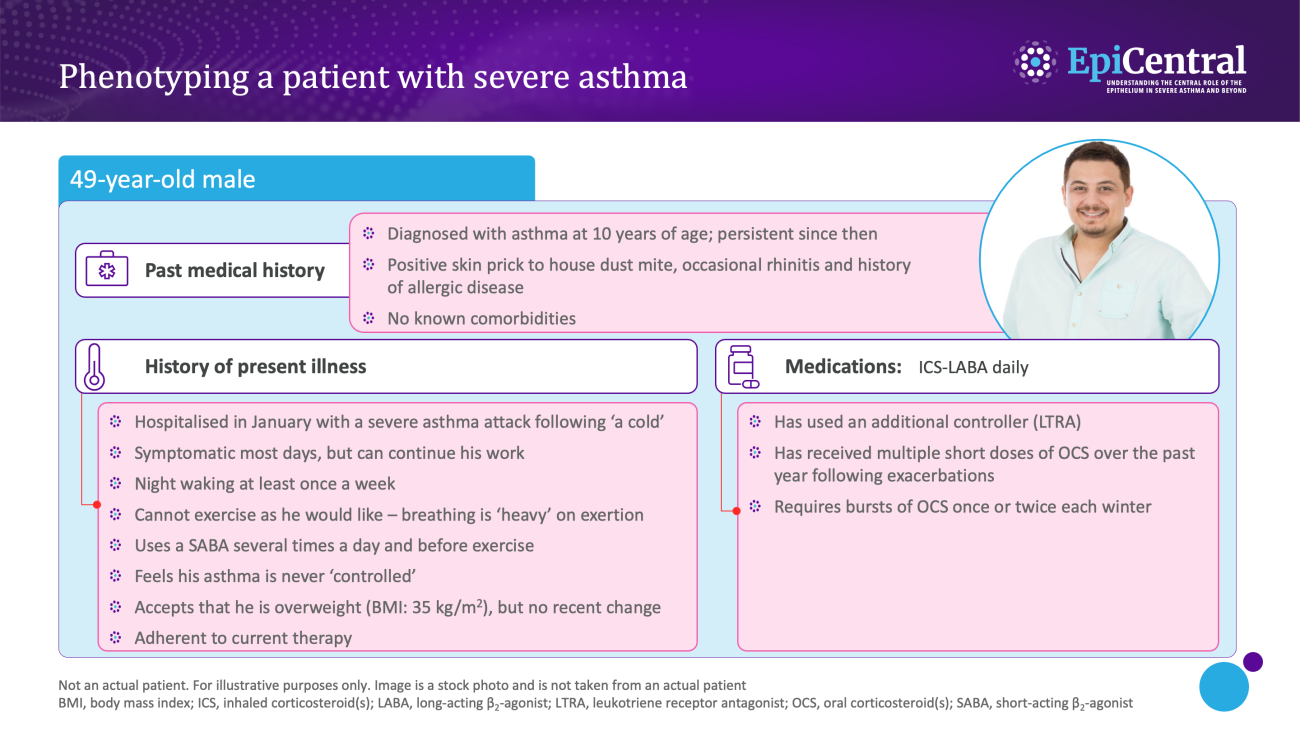
Dynamic, objective and diagnostic biomarkers can help to identify phenotypes and endotypes of severe asthma and help in the selection of an appropriate treatment.13 As such, it is important that clinicians are able to interpret biomarkers effectively to improve patient outcomes.13
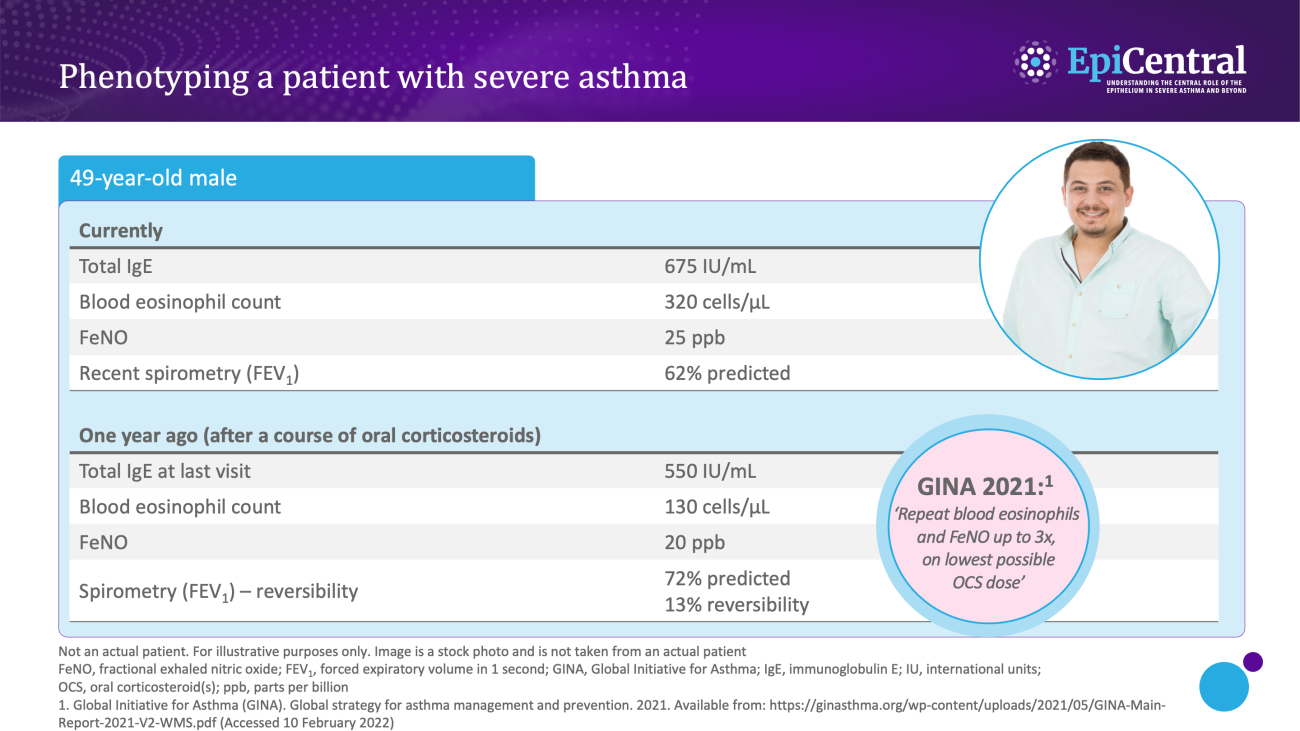
Various biomarkers of T2-mediated inflammation, including specific blood immunoglobulin (Ig) E, blood or sputum eosinophils, and fractional exhaled nitric oxide (FeNO), are available to clinicians1,13,14 and these, among others, can be used in clinical practice for phenotyping of severe asthma.13
-
Allergen-specific blood IgE levels, measured via a blood or skin prick testing, are higher in patients with allergic asthma compared with healthy individuals and can be used as a surrogate measure for atopy13,15
-
Blood eosinophils and sputum eosinophils are surrogate markers of T2-inflammation and the T2-inflammatory cytokine, interleukin (IL)-5, which is required for eosinophil activation and survival.14 These are useful biomarkers as patients with higher eosinophil counts are prone to experiencing severe disease and poorer asthma outcomes than patients with lower eosinophil counts16
-
FeNO is a biomarker of airway epithelial cell exposure to IL-13 and IL-4.10,14 These cytokines upregulate inducible nitric oxide synthase (iNOS) in the airway epithelium, and result in increased nitric oxide production.14 A high FeNO measurement correlates with airway eosinophilia in asthma and indicates increased airway T2 inflammation14